Lesson Template
The Unix Shell
The Unix shell has been around longer than most of its users have been alive. It has survived so long because it's a power tool that allows people to do complex things with just a few keystrokes. More importantly, it helps them combine existing programs in new ways and automate repetitive tasks so that they don't have to type the same things over and over again. Use of the shell is fundamental to using a wide range of other powerful tools and computing resources (including "high-performance computing" supercomputers). These lessons will start you on a path towards using these resources effectively.
Prerequisites
This lesson guides you through the basics of file systems and the shell. If you have stored files on a computer at all and recognize the word “file” and either “directory” or “folder” (two common words for the same thing), you're ready for this lesson.
If you're already comfortable manipulating files and directories, searching for files with grep and find, and writing simple loops and scripts, you probably won't learn much from this lesson.
Getting ready
You need to download some files to follow this lesson:
- Make a new folder in your Desktop called
shell-novice. - Download shell-novice-data.zip and move the file to this folder.
- If it's not unzipped yet, double-click on it to unzip it. You should end up with a new folder called
data. - You can access this folder from the Unix shell with:
$ cd && cd Desktop/shell-novice/dataTopics
- Introducing the Shell
- Files and Directories
- Creating Things
- Pipes and Filters
- Loops
- Shell Scripts
- Finding Things
Other Resources
Introducing the Shell
Learning Objectives
- Explain how the shell relates to the keyboard, the screen, the operating system, and users' programs.
- Explain when and why command-line interfaces should be used instead of graphical interfaces.
At a high level, computers do four things:
- run programs
- store data
- communicate with each other
- interact with us
They can do the last of these in many different ways, including direct brain-computer links and speech interfaces. Since these are still in their infancy, most of us use windows, icons, mice, and pointers. These technologies didn't become widespread until the 1980s, but their roots go back to Doug Engelbart's work in the 1960s, which you can see in what has been called "The Mother of All Demos".
Going back even further, the only way to interact with early computers was to rewire them. But in between, from the 1950s to the 1980s, most people used line printers. These devices only allowed input and output of the letters, numbers, and punctuation found on a standard keyboard, so programming languages and interfaces had to be designed around that constraint.
This kind of interface is called a command-line interface, or CLI, to distinguish it from a graphical user interface, or GUI, which most people now use. The heart of a CLI is a read-evaluate-print loop, or REPL: when the user types a command and then presses the enter (or return) key, the computer reads it, executes it, and prints its output. The user then types another command, and so on until the user logs off.
This description makes it sound as though the user sends commands directly to the computer, and the computer sends output directly to the user. In fact, there is usually a program in between called a command shell. What the user types goes into the shell, which then figures out what commands to run and orders the computer to execute them. Note, the shell is called the shell because it encloses the operating system in order to hide some of its complexity and make it simpler to interact with.
A shell is a program like any other. What's special about it is that its job is to run other programs rather than to do calculations itself. The most popular Unix shell is Bash, the Bourne Again SHell (so-called because it's derived from a shell written by Stephen Bourne --- this is what passes for wit among programmers). Bash is the default shell on most modern implementations of Unix and in most packages that provide Unix-like tools for Windows.
Using Bash or any other shell sometimes feels more like programming than like using a mouse. Commands are terse (often only a couple of characters long), their names are frequently cryptic, and their output is lines of text rather than something visual like a graph. On the other hand, the shell allows us to combine existing tools in powerful ways with only a few keystrokes and to set up pipelines to handle large volumes of data automatically. In addition, the command line is often the easiest way to interact with remote machines and supercomputers. Familiarity with the shell is near essential to run a variety of specialised tools and resources including high-performance computing systems. As clusters and cloud computing systems become more popular for scientific data crunching, being able to interact with them is becoming a necessary skill. We can build on the command-line skills covered here to tackle a wide range of scientific questions and computational challenges.
Nelle's Pipeline: Starting Point
Nelle Nemo, a marine biologist, has just returned from a six-month survey of the North Pacific Gyre, where she has been sampling gelatinous marine life in the Great Pacific Garbage Patch. She has 300 samples in all, and now needs to:
- Run each sample through an assay machine that will measure the relative abundance of 300 different proteins. The machine's output for a single sample is a file with one line for each protein.
- Calculate statistics for each of the proteins separately using a program her supervisor wrote called
goostat. - Compare the statistics for each protein with corresponding statistics for each other protein using a program one of the other graduate students wrote called
goodiff. - Write up results. Her supervisor would really like her to do this by the end of the month so that her paper can appear in an upcoming special issue of Aquatic Goo Letters.
It takes about half an hour for the assay machine to process each sample. The good news is that it only takes two minutes to set each one up. Since her lab has eight assay machines that she can use in parallel, this step will "only" take about two weeks.
The bad news is that if she has to run goostat and goodiff by hand, she'll have to enter filenames and click "OK" 45,150 times (300 runs of goostat, plus 300x299/2 runs of goodiff). At 30 seconds each, that will take more than two weeks. Not only would she miss her paper deadline, the chances of her typing all of those commands right are practically zero.
The next few lessons will explore what she should do instead. More specifically, they explain how she can use a command shell to automate the repetitive steps in her processing pipeline so that her computer can work 24 hours a day while she writes her paper. As a bonus, once she has put a processing pipeline together, she will be able to use it again whenever she collects more data.
Files and Directories
Learning Objectives
- Explain the similarities and differences between a file and a directory.
- Translate an absolute path into a relative path and vice versa.
- Construct absolute and relative paths that identify specific files and directories.
- Explain the steps in the shell's read-run-print cycle.
- Identify the actual command, flags, and filenames in a command-line call.
- Demonstrate the use of tab completion, and explain its advantages.
The part of the operating system responsible for managing files and directories is called the file system. It organizes our data into files, which hold information, and directories (also called "folders"), which hold files or other directories.
Several commands are frequently used to create, inspect, rename, and delete files and directories. To start exploring them, let's open a shell window:
$The dollar sign is a prompt, which shows us that the shell is waiting for input; your shell may use a different character as a prompt and may add information before the prompt. When typing commands, either from these lessons or from other sources, do not type the prompt, only the commands that follow it.
Type the command whoami, then press the Enter key (sometimes marked Return) to send the command to the shell. The command's output is the ID of the current user, i.e., it shows us who the shell thinks we are:
$ whoaminelleMore specifically, when we type whoami the shell:
- finds a program called
whoami, - runs that program,
- displays that program's output, then
- displays a new prompt to tell us that it's ready for more commands.
Next, let's find out where we are by running a command called pwd (which stands for "print working directory"). At any moment, our current working directory is our current default directory, i.e., the directory that the computer assumes we want to run commands in unless we explicitly specify something else. Here, the computer's response is /Users/nelle, which is Nelle's home directory:
$ pwd/Users/nelleTo understand what a "home directory" is, let's have a look at how the file system as a whole is organized. At the top is the root directory that holds everything else. We refer to it using a slash character / on its own; this is the leading slash in /Users/nelle.
Inside that directory are several other directories: bin (which is where some built-in programs are stored), data (for miscellaneous data files), Users (where users' personal directories are located), tmp (for temporary files that don't need to be stored long-term), and so on:
The File System
We know that our current working directory /Users/nelle is stored inside /Users because /Users is the first part of its name. Similarly, we know that /Users is stored inside the root directory / because its name begins with /.
Underneath /Users, we find one directory for each user with an account on this machine. The Mummy's files are stored in /Users/imhotep, Wolfman's in /Users/larry, and ours in /Users/nelle, which is why nelle is the last part of the directory's name.
Home Directories
Let's see what's in Nelle's home directory by running ls, which stands for "listing":
$ lscreatures molecules pizza.cfg
data north-pacific-gyre solar.pdf
Desktop notes.txt writingNelle's Home Directory
ls prints the names of the files and directories in the current directory in alphabetical order, arranged neatly into columns. We can make its output more comprehensible by using the flag -F, which tells ls to add a trailing / to the names of directories:
$ ls -Fcreatures/ molecules/ pizza.cfg
data/ north-pacific-gyre/ solar.pdf
Desktop/ notes.txt writing/Here, we can see that /Users/nelle contains six sub-directories. The names that don't have trailing slashes, like notes.txt, pizza.cfg, and solar.pdf, are plain old files. And note that there is a space between ls and -F: without it, the shell thinks we're trying to run a command called ls-F, which doesn't exist.
Now let's take a look at what's in Nelle's data directory by running ls -F data, i.e., the command ls with the arguments -F and data. The second argument --- the one without a leading dash --- tells ls that we want a listing of something other than our current working directory:
$ ls -F dataamino-acids.txt elements/ morse.txt
pdb/ planets.txt sunspot.txtThe output shows us that there are four text files and two sub-sub-directories. Organizing things hierarchically in this way helps us keep track of our work: it's possible to put hundreds of files in our home directory, just as it's possible to pile hundreds of printed papers on our desk, but it's a self-defeating strategy.
Notice, by the way that we spelled the directory name data. It doesn't have a trailing slash: that's added to directory names by ls when we use the -F flag to help us tell things apart. And it doesn't begin with a slash because it's a relative path, i.e., it tells ls how to find something from where we are, rather than from the root of the file system.
If we run ls -F /data (with a leading slash) we get a different answer, because /data is an absolute path:
$ ls -F /dataaccess.log backup/ hardware.cfg
network.cfgThe leading / tells the computer to follow the path from the root of the file system, so it always refers to exactly one directory, no matter where we are when we run the command.
What if we want to change our current working directory? Before we do this, pwd shows us that we're in /Users/nelle, and ls without any arguments shows us that directory's contents:
$ pwd/Users/nelle$ lscreatures molecules pizza.cfg
data north-pacific-gyre solar.pdf
Desktop notes.txt writingWe can use cd followed by a directory name to change our working directory. cd stands for "change directory", which is a bit misleading: the command doesn't change the directory, it changes the shell's idea of what directory we are in.
$ cd datacd doesn't print anything, but if we run pwd after it, we can see that we are now in /Users/nelle/data. If we run ls without arguments now, it lists the contents of /Users/nelle/data, because that's where we now are:
$ pwd/Users/nelle/data$ ls -Famino-acids.txt elements/ morse.txt
pdb/ planets.txt sunspot.txtWe now know how to go down the directory tree: how do we go up? We could use an absolute path:
$ cd /Users/nellebut it's almost always simpler to use cd .. to go up one level:
$ pwd/Users/nelle/data$ cd .... is a special directory name meaning "the directory containing this one", or more succinctly, the parent of the current directory. Sure enough, if we run pwd after running cd .., we're back in /Users/nelle:
$ pwd/Users/nelleThe special directory .. doesn't usually show up when we run ls. If we want to display it, we can give ls the -a flag:
$ ls -F -a./ creatures/ notes.txt
../ data/ pizza.cfg
.bash_profile molecules/ solar.pdf
Desktop/ north-pacific-gyre/ writing/-a stands for "show all"; it forces ls to show us file and directory names that begin with ., such as .. (which, if we're in /Users/nelle, refers to the /Users directory). As you can see, it also displays another special directory that's just called ., which means "the current working directory". It may seem redundant to have a name for it, but we'll see some uses for it soon. Finally, we also see a file called .bash_profile. This file usually contains settings to customize the shell (terminal). For this lesson material it does not contain any settings. There may also be similar files called .bashrc or .bash_login. The . prefix is used to prevent these configuration files from cluttering the terminal when a standard ls command is used.
Nelle's Pipeline: Organizing Files
Knowing just this much about files and directories, Nelle is ready to organize the files that the protein assay machine will create. First, she creates a directory called north-pacific-gyre (to remind herself where the data came from). Inside that, she creates a directory called 2012-07-03, which is the date she started processing the samples. She used to use names like conference-paper and revised-results, but she found them hard to understand after a couple of years. (The final straw was when she found herself creating a directory called revised-revised-results-3.)
Each of her physical samples is labelled according to her lab's convention with a unique ten-character ID, such as "NENE01729A". This is what she used in her collection log to record the location, time, depth, and other characteristics of the sample, so she decides to use it as part of each data file's name. Since the assay machine's output is plain text, she will call her files NENE01729A.txt, NENE01812A.txt, and so on. All 1520 files will go into the same directory.
If she is in her home directory, Nelle can see what files she has using the command:
$ ls north-pacific-gyre/2012-07-03/This is a lot to type, but she can let the shell do most of the work through what is called tab completion. If she types:
$ ls norand then presses tab (the tab key on her keyboard), the shell automatically completes the directory name for her:
$ ls north-pacific-gyre/If she presses tab again, Bash will add 2012-07-03/ to the command, since it's the only possible completion. Pressing tab again does nothing, since there are 1520 possibilities; pressing tab twice brings up a list of all the files, and so on. This is called tab completion, and we will see it in many other tools as we go on.
File System for Challange Questions
Relative path resolution
If pwd displays /Users/thing, what will ls ../backup display?
../backup: No such file or directory2012-12-01 2013-01-08 2013-01-272012-12-01/ 2013-01-08/ 2013-01-27/original pnas_final pnas_sub
ls reading comprehension
If pwd displays /Users/backup, and -r tells ls to display things in reverse order, what command will display:
pnas-sub/ pnas-final/ original/ls pwdls -r -Fls -r -F /Users/backup- Either #2 or #3 above, but not #1.
Default cd action
What does the command cd without a directory name do?
- It has no effect.
- It changes the working directory to
/. - It changes the working directory to the user's home directory.
- It produces an error message.
Exploring more ls arguments
What does the command ls do when used with the -s and -h arguments?
Creating Things
Learning Objectives
- Create a directory hierarchy that matches a given diagram.
- Create files in that hierarchy using an editor or by copying and renaming existing files.
- Display the contents of a directory using the command line.
- Delete specified files and/or directories.
We now know how to explore files and directories, but how do we create them in the first place? Let's go back to Nelle's home directory, /Users/nelle, and use ls -F to see what it contains:
$ pwd/Users/nelle$ ls -Fcreatures/ molecules/ pizza.cfg
data/ north-pacific-gyre/ solar.pdf
Desktop/ notes.txt writing/Let's create a new directory called thesis using the command mkdir thesis (which has no output):
$ mkdir thesisAs you might (or might not) guess from its name, mkdir means "make directory". Since thesis is a relative path (i.e., doesn't have a leading slash), the new directory is created in the current working directory:
$ ls -Fcreatures/ north-pacific-gyre/ thesis/
data/ notes.txt writing/
Desktop/ pizza.cfg
molecules/ solar.pdfHowever, there's nothing in it yet:
$ ls -F thesisLet's change our working directory to thesis using cd, then run a text editor called Nano to create a file called draft.txt:
$ cd thesis
$ nano draft.txtLet's type in a few lines of text, then use Control-O to write our data to disk:
Nano in action
Once our file is saved, we can use Control-X to quit the editor and return to the shell. (Unix documentation often uses the shorthand ^A to mean "control-A".) nano doesn't leave any output on the screen after it exits, but ls now shows that we have created a file called draft.txt:
$ lsdraft.txtLet's tidy up by running rm draft.txt:
$ rm draft.txtThis command removes files ("rm" is short for "remove"). If we run ls again, its output is empty once more, which tells us that our file is gone:
$ lsLet's re-create that file and then move up one directory to /Users/nelle using cd ..:
$ pwd/Users/nelle/thesis$ nano draft.txt
$ lsdraft.txt$ cd ..If we try to remove the entire thesis directory using rm thesis, we get an error message:
$ rm thesisrm: cannot remove `thesis': Is a directoryThis happens because rm only works on files, not directories. The right command is rmdir, which is short for "remove directory". It doesn't work yet either, though, because the directory we're trying to remove isn't empty:
$ rmdir thesisrmdir: failed to remove `thesis': Directory not emptyThis little safety feature can save you a lot of grief, particularly if you are a bad typist. To really get rid of thesis we must first delete the file draft.txt:
$ rm thesis/draft.txtThe directory is now empty, so rmdir can delete it:
$ rmdir thesisLet's create that directory and file one more time. (Note that this time we're running nano with the path thesis/draft.txt, rather than going into the thesis directory and running nano on draft.txt there.)
$ pwd/Users/nelle$ mkdir thesis$ nano thesis/draft.txt
$ ls thesisdraft.txtdraft.txt isn't a particularly informative name, so let's change the file's name using mv, which is short for "move":
$ mv thesis/draft.txt thesis/quotes.txtThe first parameter tells mv what we're "moving", while the second is where it's to go. In this case, we're moving thesis/draft.txt to thesis/quotes.txt, which has the same effect as renaming the file. Sure enough, ls shows us that thesis now contains one file called quotes.txt:
$ ls thesisquotes.txtJust for the sake of inconsistency, mv also works on directories --- there is no separate mvdir command.
Let's move quotes.txt into the current working directory. We use mv once again, but this time we'll just use the name of a directory as the second parameter to tell mv that we want to keep the filename, but put the file somewhere new. (This is why the command is called "move".) In this case, the directory name we use is the special directory name . that we mentioned earlier.
$ mv thesis/quotes.txt .The effect is to move the file from the directory it was in to the current working directory. ls now shows us that thesis is empty:
$ ls thesisFurther, ls with a filename or directory name as a parameter only lists that file or directory. We can use this to see that quotes.txt is still in our current directory:
$ ls quotes.txtquotes.txtThe cp command works very much like mv, except it copies a file instead of moving it. We can check that it did the right thing using ls with two paths as parameters --- like most Unix commands, ls can be given thousands of paths at once:
$ cp quotes.txt thesis/quotations.txt
$ ls quotes.txt thesis/quotations.txtquotes.txt thesis/quotations.txtTo prove that we made a copy, let's delete the quotes.txt file in the current directory and then run that same ls again.
$ rm quotes.txt
$ ls quotes.txt thesis/quotations.txtls: cannot access quotes.txt: No such file or directory
thesis/quotations.txtThis time it tells us that it can't find quotes.txt in the current directory, but it does find the copy in thesis that we didn't delete.
Renaming files
Suppose that you created a .txt file in your current directory to contain a list of the statistical tests you will need to do to analyze your data, and named it: statstics.txt
After creating and saving this file you realize you misspelled the filename! You want to correct the mistake, which of the following commands could you use to do so?
cp statstics.txt statistics.txtmv statstics.txt statistics.txtmv statstics.txt .cp statstics.txt .
Moving and Copying
What is the output of the closing ls command in the sequence shown below?
$ pwd
/Users/jamie/data
$ ls
proteins.dat
$ mkdir recombine
$ mv proteins.dat recombine
$ cp recombine/proteins.dat ../proteins-saved.dat
$ lsproteins-saved.dat recombinerecombineproteins.dat recombineproteins-saved.dat
Organizing Directories and Files
Jamie is working on a project and she sees that her files aren't very well organized:
$ ls -F
analyzed/ fructose.dat raw/ sucrose.datThe fructose.dat and sucrose.dat files contain output from her data analysis. What command(s) covered in this lesson does she need to run so that the commands below will produce the output shown?
$ ls -F
analyzed/ raw/
$ ls analyzed
fructose.dat sucrose.datCopy with Multiple Filenames
What does cp do when given several filenames and a directory name, as in:
$ mkdir backup
$ cp thesis/citations.txt thesis/quotations.txt backupWhat does cp do when given three or more filenames, as in:
$ ls -F
intro.txt methods.txt survey.txt
$ cp intro.txt methods.txt survey.txtListing Recursively and By Time
The command ls -R lists the contents of directories recursively, i.e., lists their sub-directories, sub-sub-directories, and so on in alphabetical order at each level. The command ls -t lists things by time of last change, with most recently changed files or directories first. In what order does ls -R -t display things?
Pipes and Filters
Learning Objectives
- Redirect a command's output to a file.
- Process a file instead of keyboard input using redirection.
- Construct command pipelines with two or more stages.
- Explain what usually happens if a program or pipeline isn't given any input to process.
- Explain Unix's "small pieces, loosely joined" philosophy.
Now that we know a few basic commands, we can finally look at the shell's most powerful feature: the ease with which it lets us combine existing programs in new ways. We'll start with a directory called molecules that contains six files describing some simple organic molecules. The .pdb extension indicates that these files are in Protein Data Bank format, a simple text format that specifies the type and position of each atom in the molecule.
$ ls moleculescubane.pdb ethane.pdb methane.pdb
octane.pdb pentane.pdb propane.pdbLet's go into that directory with cd and run the command wc *.pdb. wc is the "word count" command: it counts the number of lines, words, and characters in files. The * in *.pdb matches zero or more characters, so the shell turns *.pdb into a complete list of .pdb files:
$ cd molecules
$ wc *.pdb 20 156 1158 cubane.pdb
12 84 622 ethane.pdb
9 57 422 methane.pdb
30 246 1828 octane.pdb
21 165 1226 pentane.pdb
15 111 825 propane.pdb
107 819 6081 totalIf we run wc -l instead of just wc, the output shows only the number of lines per file:
$ wc -l *.pdb 20 cubane.pdb
12 ethane.pdb
9 methane.pdb
30 octane.pdb
21 pentane.pdb
15 propane.pdb
107 totalWe can also use -w to get only the number of words, or -c to get only the number of characters.
Which of these files is shortest? It's an easy question to answer when there are only six files, but what if there were 6000? Our first step toward a solution is to run the command:
$ wc -l *.pdb > lengths.txtThe greater than symbol, >, tells the shell to redirect the command's output to a file instead of printing it to the screen. The shell will create the file if it doesn't exist, or overwrite the contents of that file if it does. (This is why there is no screen output: everything that wc would have printed has gone into the file lengths.txt instead.) ls lengths.txt confirms that the file exists:
$ ls lengths.txtlengths.txtWe can now send the content of lengths.txt to the screen using cat lengths.txt. cat stands for "concatenate": it prints the contents of files one after another. There's only one file in this case, so cat just shows us what it contains:
$ cat lengths.txt 20 cubane.pdb
12 ethane.pdb
9 methane.pdb
30 octane.pdb
21 pentane.pdb
15 propane.pdb
107 totalNow let's use the sort command to sort its contents. We will also use the -n flag to specify that the sort is numerical instead of alphabetical. This does not change the file; instead, it sends the sorted result to the screen:
$ sort -n lengths.txt 9 methane.pdb
12 ethane.pdb
15 propane.pdb
20 cubane.pdb
21 pentane.pdb
30 octane.pdb
107 totalWe can put the sorted list of lines in another temporary file called sorted-lengths.txt by putting > sorted-lengths.txt after the command, just as we used > lengths.txt to put the output of wc into lengths.txt. Once we've done that, we can run another command called head to get the first few lines in sorted-lengths.txt:
$ sort -n lengths.txt > sorted-lengths.txt
$ head -1 sorted-lengths.txt 9 methane.pdbUsing the parameter -1 with head tells it that we only want the first line of the file; -20 would get the first 20, and so on. Since sorted-lengths.txt contains the lengths of our files ordered from least to greatest, the output of head must be the file with the fewest lines.
If you think this is confusing, you're in good company: even once you understand what wc, sort, and head do, all those intermediate files make it hard to follow what's going on. We can make it easier to understand by running sort and head together:
$ sort -n lengths.txt | head -1 9 methane.pdbThe vertical bar between the two commands is called a pipe. It tells the shell that we want to use the output of the command on the left as the input to the command on the right. The computer might create a temporary file if it needs to, or copy data from one program to the other in memory, or something else entirely; we don't have to know or care.
We can use another pipe to send the output of wc directly to sort, which then sends its output to head:
$ wc -l *.pdb | sort -n | head -1 9 methane.pdbThis is exactly like a mathematician nesting functions like log(3x) and saying "the log of three times x". In our case, the calculation is "head of sort of line count of *.pdb".
Here's what actually happens behind the scenes when we create a pipe. When a computer runs a program --- any program --- it creates a process in memory to hold the program's software and its current state. Every process has an input channel called standard input. (By this point, you may be surprised that the name is so memorable, but don't worry: most Unix programmers call it "stdin". Every process also has a default output channel called standard output (or "stdout").
The shell is actually just another program. Under normal circumstances, whatever we type on the keyboard is sent to the shell on its standard input, and whatever it produces on standard output is displayed on our screen. When we tell the shell to run a program, it creates a new process and temporarily sends whatever we type on our keyboard to that process's standard input, and whatever the process sends to standard output to the screen.
Here's what happens when we run wc -l *.pdb > lengths.txt. The shell starts by telling the computer to create a new process to run the wc program. Since we've provided some filenames as parameters, wc reads from them instead of from standard input. And since we've used > to redirect output to a file, the shell connects the process's standard output to that file.
If we run wc -l *.pdb | sort -n instead, the shell creates two processes (one for each process in the pipe) so that wc and sort run simultaneously. The standard output of wc is fed directly to the standard input of sort; since there's no redirection with >, sort's output goes to the screen. And if we run wc -l *.pdb | sort -n | head -1, we get three processes with data flowing from the files, through wc to sort, and from sort through head to the screen.
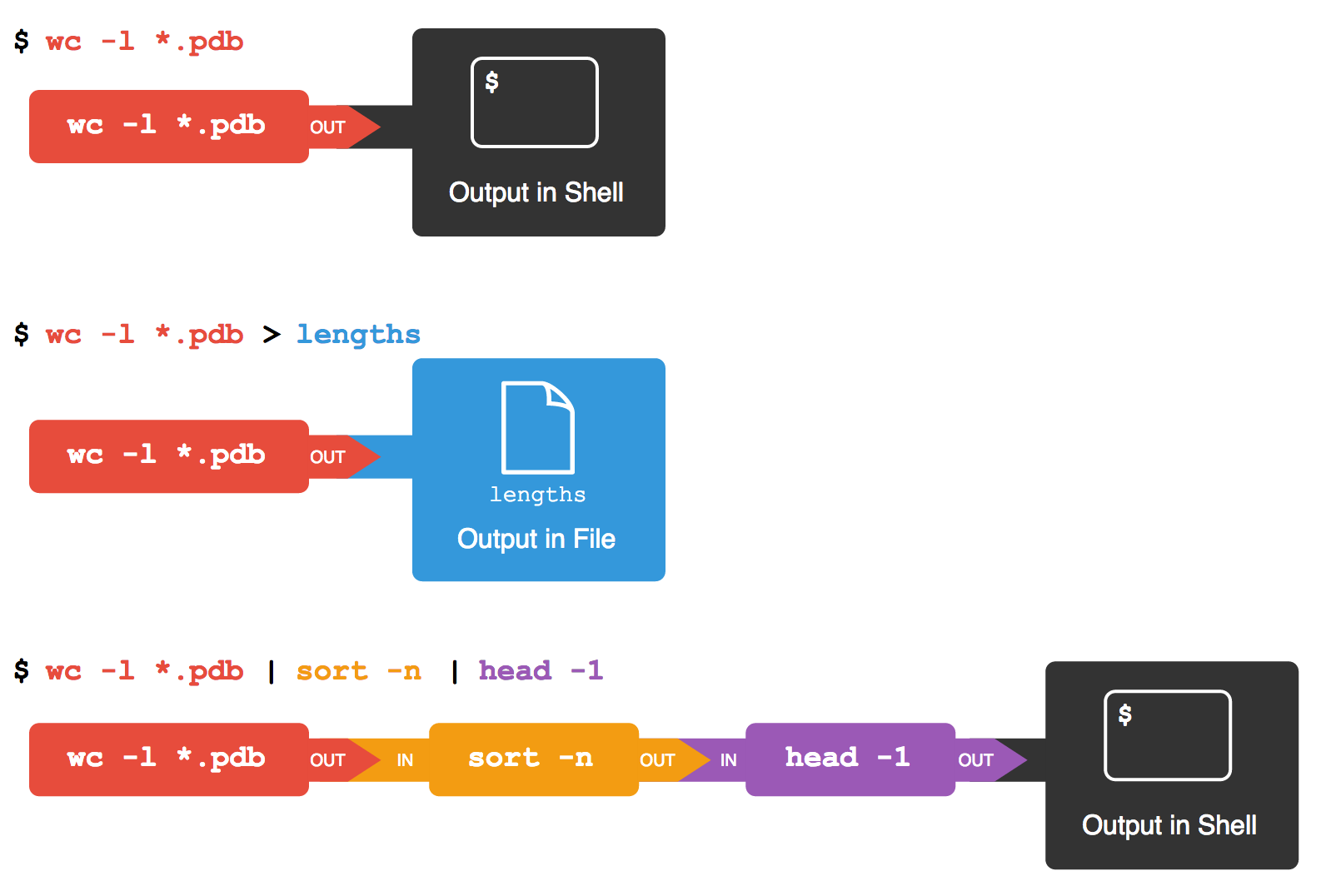
Redirects and Pipes
This simple idea is why Unix has been so successful. Instead of creating enormous programs that try to do many different things, Unix programmers focus on creating lots of simple tools that each do one job well, and that work well with each other. This programming model is called "pipes and filters". We've already seen pipes; a filter is a program like wc or sort that transforms a stream of input into a stream of output. Almost all of the standard Unix tools can work this way: unless told to do otherwise, they read from standard input, do something with what they've read, and write to standard output.
The key is that any program that reads lines of text from standard input and writes lines of text to standard output can be combined with every other program that behaves this way as well. You can and should write your programs this way so that you and other people can put those programs into pipes to multiply their power.
Nelle's Pipeline: Checking Files
Nelle has run her samples through the assay machines and created 1520 files in the north-pacific-gyre/2012-07-03 directory described earlier. As a quick sanity check, starting from her home directory, Nelle types:
$ cd north-pacific-gyre/2012-07-03
$ wc -l *.txtThe output is 1520 lines that look like this:
300 NENE01729A.txt
300 NENE01729B.txt
300 NENE01736A.txt
300 NENE01751A.txt
300 NENE01751B.txt
300 NENE01812A.txt
... ...Now she types this:
$ wc -l *.txt | sort -n | head -5 240 NENE02018B.txt
300 NENE01729A.txt
300 NENE01729B.txt
300 NENE01736A.txt
300 NENE01751A.txtWhoops: one of the files is 60 lines shorter than the others. When she goes back and checks it, she sees that she did that assay at 8:00 on a Monday morning --- someone was probably in using the machine on the weekend, and she forgot to reset it. Before re-running that sample, she checks to see if any files have too much data:
$ wc -l *.txt | sort -n | tail -5 300 NENE02040A.txt
300 NENE02040B.txt
300 NENE02040Z.txt
300 NENE02043A.txt
300 NENE02043B.txtThose numbers look good --- but what's that 'Z' doing there in the third-to-last line? All of her samples should be marked 'A' or 'B'; by convention, her lab uses 'Z' to indicate samples with missing information. To find others like it, she does this:
$ ls *Z.txtNENE01971Z.txt NENE02040Z.txtSure enough, when she checks the log on her laptop, there's no depth recorded for either of those samples. Since it's too late to get the information any other way, she must exclude those two files from her analysis. She could just delete them using rm, but there are actually some analyses she might do later where depth doesn't matter, so instead, she'll just be careful later on to select files using the wildcard expression *[AB].txt. As always, the '*' matches any number of characters; the expression [AB] matches either an 'A' or a 'B', so this matches all the valid data files she has.
What does sort -n do?
If we run sort on this file:
10
2
19
22
6the output is:
10
19
2
22
6If we run sort -n on the same input, we get this instead:
2
6
10
19
22Explain why -n has this effect.
What does < mean?
What is the difference between:
wc -l < mydata.datand:
wc -l mydata.datWhat does >> mean?
What is the difference between:
echo hello > testfile01.txtand:
echo hello >> testfile02.txtHint: Try executing each command twice in a row and then examining the output files.
Piping commands together
In our current directory, we want to find the 3 files which have the least number of lines. Which command listed below would work?
wc -l * > sort -n > head -3wc -l * | sort -n | head 1-3wc -l * | head -3 | sort -nwc -l * | sort -n | head -3
Why does uniq only remove adjacent duplicates?
The command uniq removes adjacent duplicated lines from its input. For example, if a file salmon.txt contains:
coho
coho
steelhead
coho
steelhead
steelheadthen uniq salmon.txt produces:
coho
steelhead
coho
steelheadWhy do you think uniq only removes adjacent duplicated lines? (Hint: think about very large data sets.) What other command could you combine with it in a pipe to remove all duplicated lines?
Pipe reading comprehension
A file called animals.txt contains the following data:
2012-11-05,deer
2012-11-05,rabbit
2012-11-05,raccoon
2012-11-06,rabbit
2012-11-06,deer
2012-11-06,fox
2012-11-07,rabbit
2012-11-07,bearWhat text passes through each of the pipes and the final redirect in the pipeline below?
cat animals.txt | head -5 | tail -3 | sort -r > final.txtPipe construction
The command:
$ cut -d , -f 2 animals.txtproduces the following output:
deer
rabbit
raccoon
rabbit
deer
fox
rabbit
bearWhat other command(s) could be added to this in a pipeline to find out what animals the file contains (without any duplicates in their names)?
Loops
Learning Objectives
- Write a loop that applies one or more commands separately to each file in a set of files.
- Trace the values taken on by a loop variable during execution of the loop.
- Explain the difference between a variable's name and its value.
- Explain why spaces and some punctuation characters shouldn't be used in file names.
- Demonstrate how to see what commands have recently been executed.
- Re-run recently executed commands without retyping them.
Wildcards and tab completion are two ways to reduce typing (and typing mistakes). Another is to tell the shell to do something over and over again. Suppose we have several hundred genome data files named basilisk.dat, unicorn.dat, and so on. In this example, we'll use the creatures directory which only has two example files, but the principles can be applied to many many more files at once. We would like to modify these files, but also save a version of the original files and rename them as original-basilisk.dat and original-unicorn.dat. We can't use:
$ mv *.dat original-*.datbecause that would expand to:
$ mv basilisk.dat unicorn.dat original-*.datThis wouldn't back up our files, instead we get an error:
mv: target `original-*.dat' is not a directoryThis a problem arises when mv receives more than two inputs. When this happens, it expects the last input to be a directory where it can move all the files it was passed. Since there is no directory named original-*.dat in the creatures directory we get an error.
Instead, we can use a loop to do some operation once for each thing in a list. Here's a simple example that displays the first three lines of each file in turn:
$ for filename in basilisk.dat unicorn.dat
> do
> head -3 $filename
> doneCOMMON NAME: basilisk
CLASSIFICATION: basiliscus vulgaris
UPDATED: 1745-05-02
COMMON NAME: unicorn
CLASSIFICATION: equus monoceros
UPDATED: 1738-11-24When the shell sees the keyword for, it knows it is supposed to repeat a command (or group of commands) once for each thing in a list. In this case, the list is the two filenames. Each time through the loop, the name of the thing currently being operated on is assigned to the variable called filename. Inside the loop, we get the variable's value by putting $ in front of it: $filename is basilisk.dat the first time through the loop, unicorn.dat the second, and so on.
By using the dollar sign we are telling the shell interpreter to treat filename as a variable name and substitute its value on its place, but not as some text or external command. When using variables it is also possible to put the names into curly braces to clearly delimit the variable name: $filename is equivalent to ${filename}, but is different from ${file}name. You may find this notation in other people's programs.
Finally, the command that's actually being run is our old friend head, so this loop prints out the first three lines of each data file in turn.
We have called the variable in this loop filename in order to make its purpose clearer to human readers. The shell itself doesn't care what the variable is called; if we wrote this loop as:
for x in basilisk.dat unicorn.dat
do
head -3 $x
doneor:
for temperature in basilisk.dat unicorn.dat
do
head -3 $temperature
doneit would work exactly the same way. Don't do this. Programs are only useful if people can understand them, so meaningless names (like x) or misleading names (like temperature) increase the odds that the program won't do what its readers think it does.
Here's a slightly more complicated loop:
for filename in *.dat
do
echo $filename
head -100 $filename | tail -20
doneThe shell starts by expanding *.dat to create the list of files it will process. The loop body then executes two commands for each of those files. The first, echo, just prints its command-line parameters to standard output. For example:
$ echo hello thereprints:
hello thereIn this case, since the shell expands $filename to be the name of a file, echo $filename just prints the name of the file. Note that we can't write this as:
for filename in *.dat
do
$filename
head -100 $filename | tail -20
donebecause then the first time through the loop, when $filename expanded to basilisk.dat, the shell would try to run basilisk.dat as a program. Finally, the head and tail combination selects lines 81-100 from whatever file is being processed.
Going back to our original file renaming problem, we can solve it using this loop:
for filename in *.dat
do
mv $filename original-$filename
doneThis loop runs the mv command once for each filename. The first time, when $filename expands to basilisk.dat, the shell executes:
mv basilisk.dat original-basilisk.datThe second time, the command is:
mv unicorn.dat original-unicorn.datNelle's Pipeline: Processing Files
Nelle is now ready to process her data files. Since she's still learning how to use the shell, she decides to build up the required commands in stages. Her first step is to make sure that she can select the right files --- remember, these are ones whose names end in 'A' or 'B', rather than 'Z'. Starting from her home directory, Nelle types:
$ cd north-pacific-gyre/2012-07-03
$ for datafile in *[AB].txt
> do
> echo $datafile
> doneNENE01729A.txt
NENE01729B.txt
NENE01736A.txt
...
NENE02043A.txt
NENE02043B.txtHer next step is to decide what to call the files that the goostats analysis program will create. Prefixing each input file's name with "stats" seems simple, so she modifies her loop to do that:
$ for datafile in *[AB].txt
> do
> echo $datafile stats-$datafile
> doneNENE01729A.txt stats-NENE01729A.txt
NENE01729B.txt stats-NENE01729B.txt
NENE01736A.txt stats-NENE01736A.txt
...
NENE02043A.txt stats-NENE02043A.txt
NENE02043B.txt stats-NENE02043B.txtShe hasn't actually run goostats yet, but now she's sure she can select the right files and generate the right output filenames.
Typing in commands over and over again is becoming tedious, though, and Nelle is worried about making mistakes, so instead of re-entering her loop, she presses the up arrow. In response, the shell redisplays the whole loop on one line (using semi-colons to separate the pieces):
$ for datafile in *[AB].txt; do echo $datafile stats-$datafile; doneUsing the left arrow key, Nelle backs up and changes the command echo to goostats:
$ for datafile in *[AB].txt; do bash goostats $datafile stats-$datafile; doneWhen she presses enter, the shell runs the modified command. However, nothing appears to happen --- there is no output. After a moment, Nelle realizes that since her script doesn't print anything to the screen any longer, she has no idea whether it is running, much less how quickly. She kills the job by typing Control-C, uses up-arrow to repeat the command, and edits it to read:
$ for datafile in *[AB].txt; do echo $datafile; bash goostats $datafile stats-$datafile; doneWhen she runs her program now, it produces one line of output every five seconds or so:
NENE01729A.txt
NENE01729B.txt
NENE01736A.txt
...1518 times 5 seconds, divided by 60, tells her that her script will take about two hours to run. As a final check, she opens another terminal window, goes into north-pacific-gyre/2012-07-03, and uses cat stats-NENE01729B.txt to examine one of the output files. It looks good, so she decides to get some coffee and catch up on her reading.
Variables in Loops
Suppose that ls initially displays:
fructose.dat glucose.dat sucrose.datWhat is the output of:
for datafile in *.dat
do
ls *.dat
doneNow, what is the output of:
for datafile in *.dat
do
ls $datafile
doneWhy do these two loops give you different outputs?
Saving to a File in a Loop - Part One
In the same directory, what is the effect of this loop?
for sugar in *.dat
do
echo $sugar
cat $sugar > xylose.dat
done- Prints
fructose.dat,glucose.dat, andsucrose.dat, and the text fromsucrose.datwill be saved to a file calledxylose.dat. - Prints
fructose.dat,glucose.dat, andsucrose.dat, and the text from all three files would be concatenated and saved to a file calledxylose.dat. - Prints
fructose.dat,glucose.dat,sucrose.dat, andxylose.dat, and the text fromsucrose.datwill be saved to a file calledxylose.dat. - None of the above.
Saving to a File in a Loop - Part Two
In another directory, where ls returns:
fructose.dat glucose.dat sucrose.dat maltose.txtWhat would be the output of the following loop?
for datafile in *.dat
do
cat $datafile >> sugar.dat
done- All of the text from
fructose.dat,glucose.datandsucrose.datwould be concatenated and saved to a file calledsugar.dat. - The text from
sucrose.datwill be saved to a file calledsugar.dat. - All of the text from
fructose.dat,glucose.dat,sucrose.datandmaltose.txtwould be concatenated and saved to a file calledsugar.dat. - All of the text from
fructose.dat,glucose.datandsucrose.datwould be printed to the screen and saved to a file calledsugar.dat
Doing a Dry Run
Suppose we want to preview the commands the following loop will execute without actually running those commands:
for file in *.dat
do
analyze $file > analyzed-$file
doneWhat is the difference between the the two loops below, and which one would we want to run?
# Version 1
for file in *.dat
do
echo analyze $file > analyzed-$file
done# Version 2
for file in *.dat
do
echo "analyze $file > analyzed-$file"
doneNested Loops and Command-Line Expressions
The expr does simple arithmetic using command-line parameters:
$ expr 3 + 5
8
$ expr 30 / 5 - 2
4Given this, what is the output of:
for left in 2 3
do
for right in $left
do
expr $left + $right
done
doneShell Scripts
Learning Objectives
- Write a shell script that runs a command or series of commands for a fixed set of files.
- Run a shell script from the command line.
- Write a shell script that operates on a set of files defined by the user on the command line.
- Create pipelines that include user-written shell scripts.
We are finally ready to see what makes the shell such a powerful programming environment. We are going to take the commands we repeat frequently and save them in files so that we can re-run all those operations again later by typing a single command. For historical reasons, a bunch of commands saved in a file is usually called a shell script, but make no mistake: these are actually small programs.
Let's start by going back to molecules/ and putting the following line in the file middle.sh:
$ cd molecules
$ cat middle.shhead -15 octane.pdb | tail -5This is a variation on the pipe we constructed earlier: it selects lines 11-15 of the file octane.pdb. Remember, we are not running it as a command just yet: we are putting the commands in a file.
Once we have saved the file, we can ask the shell to execute the commands it contains. Our shell is called bash, so we run the following command:
$ bash middle.shATOM 9 H 1 -4.502 0.681 0.785 1.00 0.00
ATOM 10 H 1 -5.254 -0.243 -0.537 1.00 0.00
ATOM 11 H 1 -4.357 1.252 -0.895 1.00 0.00
ATOM 12 H 1 -3.009 -0.741 -1.467 1.00 0.00
ATOM 13 H 1 -3.172 -1.337 0.206 1.00 0.00Sure enough, our script's output is exactly what we would get if we ran that pipeline directly.
What if we want to select lines from an arbitrary file? We could edit middle.sh each time to change the filename, but that would probably take longer than just retyping the command. Instead, let's edit middle.sh and replace octane.pdb with a special variable called $1:
$ cat middle.shhead -15 "$1" | tail -5Inside a shell script, $1 means "the first filename (or other parameter) on the command line". We can now run our script like this:
$ bash middle.sh octane.pdbATOM 9 H 1 -4.502 0.681 0.785 1.00 0.00
ATOM 10 H 1 -5.254 -0.243 -0.537 1.00 0.00
ATOM 11 H 1 -4.357 1.252 -0.895 1.00 0.00
ATOM 12 H 1 -3.009 -0.741 -1.467 1.00 0.00
ATOM 13 H 1 -3.172 -1.337 0.206 1.00 0.00or on a different file like this:
$ bash middle.sh pentane.pdbATOM 9 H 1 1.324 0.350 -1.332 1.00 0.00
ATOM 10 H 1 1.271 1.378 0.122 1.00 0.00
ATOM 11 H 1 -0.074 -0.384 1.288 1.00 0.00
ATOM 12 H 1 -0.048 -1.362 -0.205 1.00 0.00
ATOM 13 H 1 -1.183 0.500 -1.412 1.00 0.00We still need to edit middle.sh each time we want to adjust the range of lines, though. Let's fix that by using the special variables $2 and $3:
$ cat middle.shhead "$2" "$1" | tail "$3"$ bash middle.sh pentane.pdb -20 -5ATOM 14 H 1 -1.259 1.420 0.112 1.00 0.00
ATOM 15 H 1 -2.608 -0.407 1.130 1.00 0.00
ATOM 16 H 1 -2.540 -1.303 -0.404 1.00 0.00
ATOM 17 H 1 -3.393 0.254 -0.321 1.00 0.00
TER 18 1This works, but it may take the next person who reads middle.sh a moment to figure out what it does. We can improve our script by adding some comments at the top:
$ cat middle.sh# Select lines from the middle of a file.
# Usage: middle.sh filename -end_line -num_lines
head "$2" "$1" | tail "$3"A comment starts with a # character and runs to the end of the line. The computer ignores comments, but they're invaluable for helping people understand and use scripts.
What if we want to process many files in a single pipeline? For example, if we want to sort our .pdb files by length, we would type:
$ wc -l *.pdb | sort -nbecause wc -l lists the number of lines in the files (recall that wc stands for 'word count', adding the -l flag means 'count lines' instead) and sort -n sorts things numerically. We could put this in a file, but then it would only ever sort a list of .pdb files in the current directory. If we want to be able to get a sorted list of other kinds of files, we need a way to get all those names into the script. We can't use $1, $2, and so on because we don't know how many files there are. Instead, we use the special variable $@, which means, "All of the command-line parameters to the shell script." We also should put $@ inside double-quotes to handle the case of parameters containing spaces ("$@" is equivalent to "$1" "$2" ...) Here's an example:
$ cat sorted.shwc -l "$@" | sort -n$ bash sorted.sh *.pdb ../creatures/*.dat9 methane.pdb
12 ethane.pdb
15 propane.pdb
20 cubane.pdb
21 pentane.pdb
30 octane.pdb
163 ../creatures/basilisk.dat
163 ../creatures/unicorn.datWe have two more things to do before we're finished with our simple shell scripts. If you look at a script like:
wc -l "$@" | sort -nyou can probably puzzle out what it does. On the other hand, if you look at this script:
# List files sorted by number of lines.
wc -l "$@" | sort -nyou don't have to puzzle it out --- the comment at the top tells you what it does. A line or two of documentation like this make it much easier for other people (including your future self) to re-use your work. The only caveat is that each time you modify the script, you should check that the comment is still accurate: an explanation that sends the reader in the wrong direction is worse than none at all.
Second, suppose we have just run a series of commands that did something useful --- for example, that created a graph we'd like to use in a paper. We'd like to be able to re-create the graph later if we need to, so we want to save the commands in a file. Instead of typing them in again (and potentially getting them wrong) we can do this:
$ history | tail -4 > redo-figure-3.shThe file redo-figure-3.sh now contains:
297 bash goostats -r NENE01729B.txt stats-NENE01729B.txt
298 bash goodiff stats-NENE01729B.txt /data/validated/01729.txt > 01729-differences.txt
299 cut -d ',' -f 2-3 01729-differences.txt > 01729-time-series.txt
300 ygraph --format scatter --color bw --borders none 01729-time-series.txt figure-3.pngAfter a moment's work in an editor to remove the serial numbers on the commands, we have a completely accurate record of how we created that figure.
In practice, most people develop shell scripts by running commands at the shell prompt a few times to make sure they're doing the right thing, then saving them in a file for re-use. This style of work allows people to recycle what they discover about their data and their workflow with one call to history and a bit of editing to clean up the output and save it as a shell script.
Nelle's Pipeline: Creating a Script
An off-hand comment from her supervisor has made Nelle realize that she should have provided a couple of extra parameters to goostats when she processed her files. This might have been a disaster if she had done all the analysis by hand, but thanks to for loops, it will only take a couple of hours to re-do.
But experience has taught her that if something needs to be done twice, it will probably need to be done a third or fourth time as well. She runs the editor and writes the following:
# Calculate reduced stats for data files at J = 100 c/bp.
for datafile in "$@"
do
echo $datafile
bash goostats -J 100 -r $datafile stats-$datafile
done(The parameters -J 100 and -r are the ones her supervisor said she should have used.) She saves this in a file called do-stats.sh so that she can now re-do the first stage of her analysis by typing:
$ bash do-stats.sh *[AB].txtShe can also do this:
$ bash do-stats.sh *[AB].txt | wc -lso that the output is just the number of files processed rather than the names of the files that were processed.
One thing to note about Nelle's script is that it lets the person running it decide what files to process. She could have written it as:
# Calculate reduced stats for A and Site B data files at J = 100 c/bp.
for datafile in *[AB].txt
do
echo $datafile
bash goostats -J 100 -r $datafile stats-$datafile
doneThe advantage is that this always selects the right files: she doesn't have to remember to exclude the 'Z' files. The disadvantage is that it always selects just those files --- she can't run it on all files (including the 'Z' files), or on the 'G' or 'H' files her colleagues in Antarctica are producing, without editing the script. If she wanted to be more adventurous, she could modify her script to check for command-line parameters, and use *[AB].txt if none were provided. Of course, this introduces another tradeoff between flexibility and complexity.
Variables in shell scripts
In the molecules directory, you have a shell script called script.sh containing the following commands:
head $2 $1
tail $3 $1While you are in the molecules directory, you type the following command:
bash script.sh '*.pdb' -1 -1Which of the following outputs would you expect to see?
- All of the lines between the first and the last lines of each file ending in
*.pdbin the molecules directory - The first and the last line of each file ending in
*.pdbin the molecules directory - The first and the last line of each file in the molecules directory
- An error because of the quotes around
*.pdb
List unique species
Leah has several hundred data files, each of which is formatted like this:
2013-11-05,deer,5
2013-11-05,rabbit,22
2013-11-05,raccoon,7
2013-11-06,rabbit,19
2013-11-06,deer,2
2013-11-06,fox,1
2013-11-07,rabbit,18
2013-11-07,bear,1Write a shell script called species.sh that takes any number of filenames as command-line parameters, and uses cut, sort, and uniq to print a list of the unique species appearing in each of those files separately.
Find the longest file with a given extension
Write a shell script called longest.sh that takes the name of a directory and a filename extension as its parameters, and prints out the name of the file with the most lines in that directory with that extension. For example:
$ bash longest.sh /tmp/data pdbwould print the name of the .pdb file in /tmp/data that has the most lines.
Why record commands in the history before running them?
If you run the command:
history | tail -5 > recent.shthe last command in the file is the history command itself, i.e., the shell has added history to the command log before actually running it. In fact, the shell always adds commands to the log before running them. Why do you think it does this?
Script reading comprehension
Joel's data directory contains three files: fructose.dat, glucose.dat, and sucrose.dat. Explain what a script called example.sh would do when run as bash example.sh *.dat if it contained the following lines:
# Script 1
echo *.*
# Script 2
for filename in $1 $2 $3
do
cat $filename
done
# Script 3
echo $@.datFinding Things
Learning Objectives
- Use
grepto select lines from text files that match simple patterns. - Use
findto find files whose names match simple patterns. - Use the output of one command as the command-line parameters to another command.
- Explain what is meant by "text" and "binary" files, and why many common tools don't handle the latter well.
You can guess someone's age by how they talk about search: young people use "Google" as a verb, while crusty old Unix programmers use "grep". The word is a contraction of "global/regular expression/print", a common sequence of operations in early Unix text editors. It is also the name of a very useful command-line program.
grep finds and prints lines in files that match a pattern. For our examples, we will use a file that contains three haikus taken from a 1998 competition in Salon magazine. For this set of examples we're going to be working in the writing subdirectory:
$ cd
$ cd writing
$ cat haiku.txtThe Tao that is seen
Is not the true Tao, until
You bring fresh toner.
With searching comes loss
and the presence of absence:
"My Thesis" not found.
Yesterday it worked
Today it is not working
Software is like that.Let's find lines that contain the word "not":
$ grep not haiku.txtIs not the true Tao, until
"My Thesis" not found
Today it is not workingHere, not is the pattern we're searching for. It's pretty simple: every alphanumeric character matches against itself. After the pattern comes the name or names of the files we're searching in. The output is the three lines in the file that contain the letters "not".
Let's try a different pattern: "day".
$ grep day haiku.txtYesterday it worked
Today it is not workingThis time, two lines that include the letters "day" are outputted. However, these letters are contained within larger words. To restrict matches to lines containing the word "day" on its own, we can give grep with the -w flag. This will limit matches to word boundaries.
$ grep -w day haiku.txtIn this case, there aren't any, so grep's output is empty. Sometimes we don't want to search for a single word, but a phrase. This is also easy to do with grep by putting the phrase in quotes.
$ grep -w "is not" haiku.txtToday it is not workingWe've now seen that you don't have to have quotes around single words, but it is useful to use quotes when searching for multiple words. It also helps to make it easier to distinguish between the search term or phrase and the file being searched. We will use quotes in the remaining examples.
Another useful option is -n, which numbers the lines that match:
$ grep -n "it" haiku.txt5:With searching comes loss
9:Yesterday it worked
10:Today it is not workingHere, we can see that lines 5, 9, and 10 contain the letters "it".
We can combine options (i.e. flags) as we do with other Unix commands. For example, let's find the lines that contain the word "the". We can combine the option -w to find the lines that contain the word "the" and -n to number the lines that match:
$ grep -n -w "the" haiku.txt2:Is not the true Tao, until
6:and the presence of absence:Now we want to use the option -i to make our search case-insensitive:
$ grep -n -w -i "the" haiku.txt1:The Tao that is seen
2:Is not the true Tao, until
6:and the presence of absence:Now, we want to use the option -v to invert our search, i.e., we want to output the lines that do not contain the word "the".
$ grep -n -w -v "the" haiku.txt1:The Tao that is seen
3:You bring fresh toner.
4:
5:With searching comes loss
7:"My Thesis" not found.
8:
9:Yesterday it worked
10:Today it is not working
11:Software is like that.grep has lots of other options. To find out what they are, we can type man grep. man is the Unix "manual" command: it prints a description of a command and its options, and (if you're lucky) provides a few examples of how to use it.
To navigate through the man pages, you may use the up and down arrow keys to move line-by-line, or try the "b" and spacebar keys to skip up and down by full page. Quit the man pages by typing "q".
$ man grepGREP(1) GREP(1)
NAME
grep, egrep, fgrep - print lines matching a pattern
SYNOPSIS
grep [OPTIONS] PATTERN [FILE...]
grep [OPTIONS] [-e PATTERN | -f FILE] [FILE...]
DESCRIPTION
grep searches the named input FILEs (or standard input if no files are named, or if a single hyphen-
minus (-) is given as file name) for lines containing a match to the given PATTERN. By default, grep
prints the matching lines.
... ... ...
OPTIONS
Generic Program Information
--help Print a usage message briefly summarizing these command-line options and the bug-reporting
address, then exit.
-V, --version
Print the version number of grep to the standard output stream. This version number should be
included in all bug reports (see below).
Matcher Selection
-E, --extended-regexp
Interpret PATTERN as an extended regular expression (ERE, see below). (-E is specified by
POSIX.)
-F, --fixed-strings
Interpret PATTERN as a list of fixed strings, separated by newlines, any of which is to be
matched. (-F is specified by POSIX.)
... ... ...While grep finds lines in files, the find command finds files themselves. Again, it has a lot of options; to show how the simplest ones work, we'll use the directory tree shown below.
File Tree for Find Example
Nelle's writing directory contains one file called haiku.txt and four subdirectories: thesis (which is sadly empty), data (which contains two files one.txt and two.txt), a tools directory that contains the programs format and stats, and an empty subdirectory called old.
For our first command, let's run find . -type d. As always, the . on its own means the current working directory, which is where we want our search to start; -type d means "things that are directories". Sure enough, find's output is the names of the five directories in our little tree (including .):
$ find . -type d./
./data
./thesis
./tools
./tools/oldIf we change -type d to -type f, we get a listing of all the files instead:
$ find . -type f./haiku.txt
./tools/stats
./tools/old/oldtool
./tools/format
./thesis/empty-draft.md
./data/one.txt
./data/two.txtfind automatically goes into subdirectories, their subdirectories, and so on to find everything that matches the pattern we've given it. If we don't want it to, we can use -maxdepth to restrict the depth of search:
$ find . -maxdepth 1 -type f./haiku.txtThe opposite of -maxdepth is -mindepth, which tells find to only report things that are at or below a certain depth. -mindepth 2 therefore finds all the files that are two or more levels below us:
$ find . -mindepth 2 -type f./data/one.txt
./data/two.txt
./tools/format
./tools/statsNow let's try matching by name:
$ find . -name *.txt./haiku.txtWe expected it to find all the text files, but it only prints out ./haiku.txt. The problem is that the shell expands wildcard characters like * before commands run. Since *.txt in the current directory expands to haiku.txt, the command we actually ran was:
$ find . -name haiku.txtfind did what we asked; we just asked for the wrong thing.
To get what we want, let's do what we did with grep: put *.txt in single quotes to prevent the shell from expanding the * wildcard. This way, find actually gets the pattern *.txt, not the expanded filename haiku.txt:
$ find . -name '*.txt'./data/one.txt
./data/two.txt
./haiku.txtAs we said earlier, the command line's power lies in combining tools. We've seen how to do that with pipes; let's look at another technique. As we just saw, find . -name '*.txt' gives us a list of all text files in or below the current directory. How can we combine that with wc -l to count the lines in all those files?
The simplest way is to put the find command inside $():
$ wc -l $(find . -name '*.txt')11 ./haiku.txt
300 ./data/two.txt
70 ./data/one.txt
381 totalWhen the shell executes this command, the first thing it does is run whatever is inside the $(). It then replaces the $() expression with that command's output. Since the output of find is the three filenames ./data/one.txt, ./data/two.txt, and ./haiku.txt, the shell constructs the command:
$ wc -l ./data/one.txt ./data/two.txt ./haiku.txtwhich is what we wanted. This expansion is exactly what the shell does when it expands wildcards like * and ?, but lets us use any command we want as our own "wildcard".
It's very common to use find and grep together. The first finds files that match a pattern; the second looks for lines inside those files that match another pattern. Here, for example, we can find PDB files that contain iron atoms by looking for the string "FE" in all the .pdb files above the current directory:
$ grep "FE" $(find .. -name '*.pdb')../data/pdb/heme.pdb:ATOM 25 FE 1 -0.924 0.535 -0.518The Unix shell is older than most of the people who use it. It has survived so long because it is one of the most productive programming environments ever created --- maybe even the most productive. Its syntax may be cryptic, but people who have mastered it can experiment with different commands interactively, then use what they have learned to automate their work. Graphical user interfaces may be better at the first, but the shell is still unbeaten at the second. And as Alfred North Whitehead wrote in 1911, "Civilization advances by extending the number of important operations which we can perform without thinking about them."
Using grep
The Tao that is seen
Is not the true Tao, until
You bring fresh toner.
With searching comes loss
and the presence of absence:
"My Thesis" not found.
Yesterday it worked
Today it is not working
Software is like that.From the above text, contained in the file haiku.txt, which command would result in the following output:
and the presence of absence:grep "of" haiku.txtgrep -E "of" haiku.txtgrep -w "of" haiku.txtgrep -i "of" haiku.txt
find pipeline reading comprehension
Write a short explanatory comment for the following shell script:
find . -name '*.dat' | wc -l | sort -nMatching
ose.datbut nottempThe
-vflag togrepinverts pattern matching, so that only lines which do not match the pattern are printed. Given that, which of the following commands will find all files in/datawhose names end inose.dat(e.g.,sucrose.datormaltose.dat), but do not contain the wordtemp?
find /data -name '*.dat' | grep ose | grep -v temp
find /data -name ose.dat | grep -v temp
grep -v "temp" $(find /data -name '*ose.dat')None of the above.
Little Women
You and your friend, having just finished reading Little Women by Louisa May Alcott, are in an argument. Of the four sisters in the book, Jo, Meg, Beth, and Amy, your friend thinks that Jo was the most mentioned. You, however, are certain it was Amy. Luckily, you have a file
LittleWomen.txtcontaining the full text of the novel. Using aforloop, how would you tabulate the number of times each of the four sisters is mentioned? Hint: one solution might employ the commandsgrepandwcand a|, while another might utilizegrepoptions.
Reference
Introducing the Shell
- A shell is a program whose primary purpose is to read commands and run other programs.
- The shell's main advantages are its high action-to-keystroke ratio, its support for automating repetitive tasks, and that it can be used to access networked machines.
- The shell's main disadvantages are its primarily textual nature and how cryptic its commands and operation can be.
Files and Directories
- The file system is responsible for managing information on the disk.
- Information is stored in files, which are stored in directories (folders).
- Directories can also store other directories, which forms a directory tree.
cd pathchanges the current working directory.ls pathprints a listing of a specific file or directory;lson its own lists the current working directory.pwdprints the user's current working directory.whoamishows the user's current identity./on its own is the root directory of the whole file system.- A relative path specifies a location starting from the current location.
- An absolute path specifies a location from the root of the file system.
- Directory names in a path are separated with '/' on Unix, but '\' on Windows.
- '..' means "the directory above the current one"; '.' on its own means "the current directory".
- Most files' names are
something.extension. The extension isn't required, and doesn't guarantee anything, but is normally used to indicate the type of data in the file. - Most commands take options (flags) which begin with a '-'.
Creating Things
cp old newcopies a file.mkdir pathcreates a new directory.mv old newmoves (renames) a file or directory.rm pathremoves (deletes) a file.rmdir pathremoves (deletes) an empty directory.- Unix documentation uses '^A' to mean "control-A".
- The shell does not have a trash bin: once something is deleted, it's really gone.
- Nano is a very simple text editor --- please use something else for real work.
Pipes and Filters
catdisplays the contents of its inputs.headdisplays the first few lines of its input.taildisplays the last few lines of its input.sortsorts its inputs.wccounts lines, words, and characters in its inputs.command > fileredirects a command's output to a file.first | secondis a pipeline: the output of the first command is used as the input to the second.- The best way to use the shell is to use pipes to combine simple single-purpose programs (filters).
Loops
- A
forloop repeats commands once for every thing in a list. - Every
forloop needs a variable to refer to the current "thing". - Use
$nameto expand a variable (i.e., get its value). - Do not use spaces, quotes, or wildcard characters such as '*' or '?' in filenames, as it complicates variable expansion.
- Give files consistent names that are easy to match with wildcard patterns to make it easy to select them for looping.
- Use the up-arrow key to scroll up through previous commands to edit and repeat them.
- Use "control-r" to search through the previously entered commands.
- Use
historyto display recent commands, and!numberto repeat a command by number.
Shell Scripts
- Save commands in files (usually called shell scripts) for re-use.
bash filenameruns the commands saved in a file.$*refers to all of a shell script's command-line parameters.$1,$2, etc., refer to specified command-line parameters.- Place variables in quotes if the values might have spaces in them.
- Letting users decide what files to process is more flexible and more consistent with built-in Unix commands.
Finding Things
findfinds files with specific properties that match patterns.grepselects lines in files that match patterns.man commanddisplays the manual page for a given command.*matches zero or more characters in a filename, so*.txtmatches all files ending in.txt.?matches any single character in a filename, so?.txtmatchesa.txtbut notany.txt.$(command)inserts a command's output in place.man commanddisplays the manual page for a given command.
Glossary
- absolute path
- A path that refers to a particular location in a file system. Absolute paths are usually written with respect to the file system's root directory, and begin with either "/" (on Unix) or "\" (on Microsoft Windows). See also: relative path.
- argument
- A value given to a function or program when it runs. The term is often used interchangeably (and inconsistently) with parameter.
- command shell
- See shell
- command-line interface
- An interface based on typing commands, usually at a REPL. See also: graphical user interface.
- comment
- A remark in a program that is intended to help human readers understand what is going on, but is ignored by the computer. Comments in Python, R, and the Unix shell start with a
#character and run to the end of the line; comments in SQL start with--, and other languages have other conventions. - current working directory
- The directory that relative paths are calculated from; equivalently, the place where files referenced by name only are searched for. Every process has a current working directory. The current working directory is usually referred to using the shorthand notation
.(pronounced "dot"). - file system
- A set of files, directories, and I/O devices (such as keyboards and screens). A file system may be spread across many physical devices, or many file systems may be stored on a single physical device; the operating system manages access.
- filename extension
- The portion of a file's name that comes after the final "." character. By convention this identifies the file's type:
.txtmeans "text file",.pngmeans "Portable Network Graphics file", and so on. These conventions are not enforced by most operating systems: it is perfectly possible to name an MP3 sound filehomepage.html. Since many applications use filename extensions to identify the MIME type of the file, misnaming files may cause those applications to fail. - filter
- A program that transforms a stream of data. Many Unix command-line tools are written as filters: they read data from standard input, process it, and write the result to standard output.
- flag
- A terse way to specify an option or setting to a command-line program. By convention Unix applications use a dash followed by a single letter, such as
-v, or two dashes followed by a word, such as--verbose, while DOS applications use a slash, such as/V. Depending on the application, a flag may be followed by a single argument, as in-o /tmp/output.txt. - for loop
- A loop that is executed once for each value in some kind of set, list, or range. See also: while loop.
- graphical user interface
- A graphical user interface, usually controlled by using a mouse. See also: command-line interface.
- home directory
- The default directory associated with an account on a computer system. By convention, all of a user's files are stored in or below her home directory.
- loop
- A set of instructions to be executed multiple times. Consists of a loop body and (usually) a condition for exiting the loop. See also for loop and while loop.
- loop body
- The set of statements or commands that are repeated inside a for loop or while loop.
- MIME type
- MIME (Multi-Purpose Internet Mail Extensions) types describe different file types for exchange on the Internet, for example images, audio, and documents.
- operating system
- Software that manages interactions between users, hardware, and software processes. Common examples are Linux, OS X, and Windows.
- orthogonal
- To have meanings or behaviors that are independent of each other. If a set of concepts or tools are orthogonal, they can be combined in any way.
- parameter
- A variable named in the function's declaration that is used to hold a value passed into the call. The term is often used interchangeably (and inconsistently) with argument.
- parent directory
- The directory that "contains" the one in question. Every directory in a file system except the root directory has a parent. A directory's parent is usually referred to using the shorthand notation
..(pronounced "dot dot"). - path
- A description that specifies the location of a file or directory within a file system. See also: absolute path, relative path.
- pipe
- A connection from the output of one program to the input of another. When two or more programs are connected in this way, they are called a "pipeline".
- process
- A running instance of a program, containing code, variable values, open files and network connections, and so on. Processes are the "actors" that the operating system manages; it typically runs each process for a few milliseconds at a time to give the impression that they are executing simultaneously.
- prompt
- A character or characters display by a REPL to show that it is waiting for its next command.
- quoting
- (in the shell): Using quotation marks of various kinds to prevent the shell from interpreting special characters. For example, to pass the string
*.txtto a program, it is usually necessary to write it as'*.txt'(with single quotes) so that the shell will not try to expand the*wildcard. - read-evaluate-print loop
- (REPL): A command-line interface that reads a command from the user, executes it, prints the result, and waits for another command.
- redirect
- To send a command's output to a file rather than to the screen or another command, or equivalently to read a command's input from a file.
- regular expression
- A pattern that specifies a set of character strings. REs are most often used to find sequences of characters in strings.
- relative path
- A path that specifies the location of a file or directory with respect to the current working directory. Any path that does not begin with a separator character ("/" or "\") is a relative path. See also: absolute path.
- root directory
- The top-most directory in a file system. Its name is "/" on Unix (including Linux and Mac OS X) and "\" on Microsoft Windows.
- shell
- A command-line interface such as Bash (the Bourne-Again Shell) or the Microsoft Windows DOS shell that allows a user to interact with the operating system.
- shell script
- A set of shell commands stored in a file for re-use. A shell script is a program executed by the shell; the name "script" is used for historical reasons.
- standard input
- A process's default input stream. In interactive command-line applications, it is typically connected to the keyboard; in a pipe, it receives data from the standard output of the preceding process.
- standard output
- A process's default output stream. In interactive command-line applications, data sent to standard output is displayed on the screen; in a pipe, it is passed to the standard input of the next process.
- sub-directory
- A directory contained within another directory.
- tab completion
- A feature provided by many interactive systems in which pressing the Tab key triggers automatic completion of the current word or command.
- variable
- A name in a program that is associated with a value or a collection of values.
- while loop
- A loop that keeps executing as long as some condition is true. See also: for loop.
- wildcard
- A character used in pattern matching. In the Unix shell, the wildcard
*matches zero or more characters, so that*.txtmatches all files whose names end in.txt.
Discussion
Job Control Codes
The shell accepts a few special commands that allow users to interact with running processes or programs. You can enter each of these "control codes" by holding down the CTRL key and then pressing one of the control characters. In other tutorials, you may see the ^ character used to represent the CTRL key (e.g. ^c = CTRL+c).
| Control Code | Description |
|---|---|
| CTRL+c | Interrupts and cancels a running program. This is useful if you want to cancel a command that is taking too long to execute. |
| CTRL+d | Indicates the end of a file or stream of characters that you are entering on the command line. For example, we saw earlier that the wc command counts lines, words, and characters in a file. If we just type wc and hit Enter without providing a file name, then wc will assume we want it to analyze all the stuff we type next. After typing our magnum opus directly into the shell prompt, we can then type CTRL+d to tell wc that we're done and we'd like to see the results of the word count. |
| CTRL+z | Suspends a process but does not terminate it. You can then use the command fg to restart the job in the foreground. |
For new shell users, these control codes can all appear to have the same effect: they make things "go away." But it is helpful to understand the differences. In general, if something went wrong and you just want to get your shell prompt back, it is better to use CTRL+c.
Other Shells
Before Bash became popular in the end of nineties, scientists widely used (and some still use) another shell, C-shell, or Csh. Bash and Csh have similar feature sets, but their syntax rules are different and this makes them incompatible with each other. A few other shells have appeared since, including ksh, zsh, and a number of others; they are mostly compatible with Bash, and Bash is the default shell on most modern implementations of Unix (including most packages that provide Unix-like tools for Windows) but if you get strange errors in shell scripts written by colleagues, check to see which shell they were written for.
Instructor's Guide
- Why do we learn to use the shell?
- Allows users to automate repetitive tasks
- And capture small data manipulation steps that are normally not recorded to make research reproducible
- The Problem
- Running the same workflow on several samples can be unnecessarily labour intensive
- Manual manipulation of data files:
- is often not captured in documentation
- is hard to reproduce
- is hard to troubleshoot, review, or improve
- The Shell
- Workflows can be automated through the use of shell scripts
- Built-in commands allow for easy data manipulation (e.g. sort, grep, etc.)
- Every step can be captured in the shell script and allow reproducibility and easy troubleshooting
Overall
Many people have questioned whether we should still teach the shell. After all, anyone who wants to rename several thousand data files can easily do so interactively in the Python interpreter, and anyone who's doing serious data analysis is probably going to do most of their work inside the IPython Notebook or R Studio. So why teach the shell?
The first answer is, "Because so much else depends on it." Installing software, configuring your default editor, and controlling remote machines frequently assume a basic familiarity with the shell, and with related ideas like standard input and output. Many tools also use its terminology (for example, the %ls and %cd magic commands in IPython).
The second answer is, "Because it's an easy way to introduce some fundamental ideas about how to use computers." As we teach people how to use the Unix shell, we teach them that they should get the computer to repeat things (via tab completion, ! followed by a command number, and for loops) rather than repeating things themselves. We also teach them to take things they've discovered they do frequently and save them for later re-use (via shell scripts), to give things sensible names, and to write a little bit of documentation (like comment at the top of shell scripts) to make their future selves' lives better.
The third answer is, "Because it enables use of many domain-specific tools and compute resources researchers cannot access otherwise." Familiarity with the shell is very useful for remote accessing machines, using high-performance computing infrastructure, and running new specialist tools in many disciplines. We do not teach HPC or domain-specific skills here but lay the groundwork for further development of these skills. In particular, understanding the syntax of commands, flags, and help systems is useful for domain specific tools and understanding the file system (and how to navigate it) is useful for remote access.
Finally, and perhaps most importantly, teaching people the shell lets us teach them to think about programming in terms of function composition. In the case of the shell, this takes the form of pipelines rather than nested function calls, but the core idea of "small pieces, loosely joined" is the same.
All of this material can be covered in three hours as long as learners using Windows do not run into roadblocks such as:
- not being able to figure out where their home directory is (particularly if they're using Cygwin);
- not being able to run a plain text editor; and
- the shell refusing to run scripts that include DOS line endings.
Teaching Notes
- Time estimates:
- @gvwilson: 3 hours
Super cool online resource! http://explainshell.com/ will dissect any shell command you type in and display help text for each piece.
Resources for "splitting" your shell so that recent commands remain in view: http://blog.rgaiacs.com/2015/06/18/swc_shell.html
- How to use the materials in the shell-novice repository (or, lesson planning)
- For a great general list of tips, see this swcarpentry blog post
- Use the
datadirectory for in-workshop exercises and live coding examples. You can clone the shell-novice directory or use theDownload ZIPbutton on the right to get the entire repository; we also now provide a zip file of thedatadirectory that can be downloaded on its own from the repository by right-click + save. See the "Get Ready" box on the front page of the website for more details.
- Website: various practices have been used.
- Option 1: Can give links to learners before the lesson so they can follow along, catch up, and see exercises (particularly if you're following the lesson content without many changes).
- Option 2: Don't show the website to the learners during the lesson, as it can be distracting - students may read instead of listen, and having another window open is an additional cognitive load.
- In any case, make sure to point to website as a post-workshop reference.
- Content: Unless you have a truly generous amount of time (4+ hours), it is likely that you will not cover ALL the material in this lesson in a single half-day session. Plan ahead on what you might skip, what you really want to emphasize, etc.
- Exercises: Think in advance about how you might want to handle exercises during the lesson. How are you assigning them (website, slide, handout)? Do you want everyone to try it and then you show the solution? Have a learner show the solution? Have groups each do a different exercise and present their solutions?
reference.mdcan be printed out and given to students as a reference, your choice.- Other preparation: Feel free to add your own examples or side comments, but know that it shouldn't be necessary - the topics and commands can be taught as given on the lesson pages! If you think there is a place where the lesson is lacking, feel free to raise an issue or submit a pull request.
- Setup:
- Run
tools/gen-nene.pyto regenerate random data files if needed (some are already in thefilesystemdirectory). - Run
tools/gen-sequence.pyto regenerate random sequence data if needed.
- Run
Have learners open a shell and then do
whoami,pwd, andls. Then have them create a directory calledworkshopandcdinto it, so that everything else they do during the lesson is unlikely to harm whatever files they already have.Get them to run an editor and save a file in their
workshopdirectory as early as possible. Doing this is usually the biggest stumbling block during the entire lesson: many will try to run the same editor as the instructor (which may leave them trapped in the awful nether hell that is Vim), or will not know how to navigate to the right directory to save their file, or will run a word processor rather than a plain text editor. The quickest way past these problems is to have more knowledgeable learners help those who need it.Tab completion sounds like a small thing: it isn't. Re-running old commands using
!123or!wcisn't a small thing either, and neither are wildcard expansion andforloops. Each one is an opportunity to repeat one of the big ideas of Software Carpentry: if the computer can repeat it, some programmer somewhere will almost certainly have built some way for the computer to repeat it.Building up a pipeline with four or five stages, then putting it in a shell script for re-use and calling that script inside a
forloop, is a great opportunity to show how "seven plus or minus two" connects to programming. Once we have figured out how to do something moderately complicated, we make it re-usable and give it a name so that it only takes up one slot in working memory rather than several. It is also a good opportunity to talk about exploratory programming: rather than designing a program up front, we can do a few useful things and then retroactively decide which are worth encapsulating for future re-use.If everything is going well, you can drive home the point that file extensions are essentially there to help computers (and human readers) understand file content and are not a requirement of files (covered briefly in Files and Directories). This can be done in the Pipes and Filters section by showing that you can redirect standard output to a file without the .txt extension (e.g., lengths), and that the resulting file is still a perfectly usable text file. Make the point that if double-clicked in the GUI, the computer will probably ask you what you want to do.
We have to leave out many important things because of time constraints, including file permissions, job control, and SSH. If learners already understand the basic material, this can be covered instead using the online lessons as guidelines. These limitations also have follow-on consequences:
It's hard to discuss
#!(shebang) without first discussing permissions, which we don't do.Installing Bash and a reasonable set of Unix commands on Windows always involves some fiddling and frustration. Please see the latest set of installation guidelines for advice, and try it out yourself before teaching a class.
On Windows, it appears that:
$ cd $ cd Desktopwill always put someone on their desktop. Have them create the example directory for the shell exercises there so that they can find it easily and watch it evolve.
Stay within POSIX-compliant commands, as all the teaching materials do. Your particular shell may have extensions beyond POSIX that are not available on other machines, especially the default OSX bash and Windows bash emulators. For example, POSIX
lsdoes not have an--ignore=or-Ioption, and POSIXheadtakes-n 10or-10, but not the long form of--lines=10.
Windows
Installing Bash and a reasonable set of Unix commands on Windows always involves some fiddling and frustration. Please see the latest set of installation guidelines for advice, and try it out yourself before teaching a class. Options we have explored include:
- msysGit (also called "Git Bash"),
- Cygwin,
- using a desktop virtual machine, and
- having learners connect to a remote Unix machine (typically a VM in the cloud).
Cygwin was the preferred option until mid-2013, but once we started teaching Git, msysGit proved to work better. Desktop virtual machines and cloud-based VMs work well for technically sophisticated learners, and can reduce installation and configuration at the start of the workshop, but:
- they don't work well on underpowered machines,
- they're confusing for novices (because simple things like copy and paste work differently),
- learners leave the workshop without a working environment on their operating system of choice, and
- learners may show up without having downloaded the VM or the wireless will go down (or become congested) during the lesson.
Whatever you use, please test it yourself on a Windows machine before your workshop: things may always have changed behind your back since your last workshop. And please also make use of our Windows setup helper.
Licenses
Instructional Material
All Software Carpentry instructional material is made available under the Creative Commons Attribution license. The following is a human-readable summary of (and not a substitute for) the full legal text of the CC BY 4.0 license.
You are free:
- to Share---copy and redistribute the material in any medium or format
- to Adapt---remix, transform, and build upon the material
for any purpose, even commercially.
The licensor cannot revoke these freedoms as long as you follow the license terms.
Under the following terms:
- Attribution---You must give appropriate credit (mentioning that your work is derived from work that is Copyright © Software Carpentry and, where practical, linking to http://software-carpentry.org/), provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
No additional restrictions---You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits. With the understanding that:
Notices:
- You do not have to comply with the license for elements of the material in the public domain or where your use is permitted by an applicable exception or limitation.
- No warranties are given. The license may not give you all of the permissions necessary for your intended use. For example, other rights such as publicity, privacy, or moral rights may limit how you use the material.
Software
Except where otherwise noted, the example programs and other software provided by Software Carpentry are made available under the OSI-approved MIT license.
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Trademark
"Software Carpentry" and the Software Carpentry logo are registered trademarks of NumFOCUS.
